Measuring the Effects of Interferon Stimulated Genes on Viral Replication
Whenever I read a paper that describes an experimental technique that I’m not familiar with (which is often, since the lab bench is not where bioinformatic students spend most their time), I compensate by doing background research. Lately, I have been reading a lot about the antiviral effects of interferon-stimulated genes (ISGs), and since John Schoggins is giving a seminar in my university on Monday, I thought I would post about a clever technique that he used in his Nature paper to quantify the impact of ISGs on viral replication.
Interferon-stimulated genes are known to inhibit viral replication, but this effect hasn’t been quantified for all of them. The goal of the Schoggins paper was to determine how individual ISGs change the ability of different viruses to replicate.
The authors used a bicistronic lentiviral vector, which is molecular biology jargon for a piece of DNA that contains two genes that get inserted into the genome of a cell by using a lentivirus virion. They are also known as plasmids or constructs. Besides the two genes, the foreign DNA also contains a bunch of other interesting molecular tools assembled in Frankenstein-like fashion:

LTR stands for long terminal repeat. It is a sequence of nucleotides repeated hundreds of times. Viruses (especially retroviruses, like HIV) use them to insert their genetic material into the genome of their target cell. In this case, instead of inserting a viral genome, they help integrate the plasmid into the genome of a cell.
CMV is one of the enhancer sequences of the human cytomegalovirus genome. It is another viral tool to increase the amount of gene transcription.
IVS-beta is the second intron in the rabbit beta-globin gene (one of the chains that makes up hemoglobin). Adding it to the construct also increases the transcription of the plasmid genes. I haven’t been able to really understand why, but I think it has to do with how it interacts with the splicing machinery.
ISG is the sequence for one of the 300+ known interferon-stimulated genes. Inserting it after the two previous elements ensures that it will be highly transcribed.
EMCV IRES is the internal ribosomal entry site of the encephalomyocarditis virus. This is another page taken from the virus book: it is a nucleotide sequence that allows ribosomes to bind to the middle of the messenger RNA molecule, instead of the more traditional 5’ end. Viruses use it to synthesize their proteins when the host translational machinery is not supposed to be working. In the plasmid above, it ensures that the RFP gene is translated in a proportional amount to the ISG gene.
RFP stands for red fluorescent protein. Using it as the second protein of the plasmid makes it easy to identify which cells have incorporated the plasmid into their genome, and are therefore, also expressing the ISG of interest.
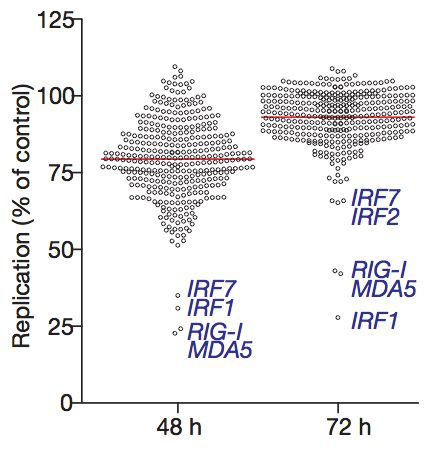
The viruses used in the experiment were modified so they would express GFP (green fluorescent protein). That way, the amount of viral replication can be quantified by measuring the amount of red fluorescence (from cells that incorporated the plasmid) and green fluorescence (from cells that were also infected with the virus). Each ISG construct has a specific ability to reduce viral replication. Surprisingly, a number of ISGs actually enhance viral replication (the circles that are above 100% in the graph below), although the reasons for this are not yet clear.
Clever molecular techniques like this one are the best tool that we have to understand what is really going on inside our cells. By studying them, I hope to learn enough to eventually design my own useful experiments.