Tag: 'science'
I went to get a haircut the other day and my hairdresser convinced me to buy a fancy pack conditioner, which, she swore, was a thousand times better than your everyday conditioner, although she didn’t know why. After forking out the money, I walked back home fashionably stimulated but scientifically unsatisfied, and I decided to try to decipher the ingredients list from my latest acquisition and understand what made them so special.
After battling with the unpronounceable names that the International Nomenclature of Cosmetic Ingredients (or INCI) choses for their chemicals, I found that my pack conditioner had many more emulsifying chemicals than a regular conditioner.
An emulsifier is a type of surfactant, its molecules encourage the stable mixing of two liquids that wouldn’t normally mix. Mayonnaise, for example, is an emulsion of oil and water (egg yolk is 50% water), and lecithin is a fatty component found in egg yolk that acts as the emulsifier. The more emulsifiers you add to a conditioner the more strongly they bind to keratin (the protein that makes up hair). The emulsifying molecules that don’t come off after rinsing cover the hair surface and make it look healthier and less dry.
While I was knee-deep researching emulsions, Wikipedia pointed me to the Tyndall effect, which explains why shorter-wavelength light is more prone to scattering by small particles than longer-wavelength light; and by extension, why blue eyes don’t actually have any blue pigment.
Human irises without melanin are translucent—in effect, an “emulsion” between a translucent layer and embedded small particles that make it look turbid. When light particles enter the turbid layer, those that have longer wavelengths (which we perceive as red) get through, and the ones with shorter wavelength (which we perceive as blue) get back-scattered by the small particles embedded in the iris. We know that wavelength affects electromagnetic radiation from our everyday experience: radio waves (longer wavelengths) can traverse walls so you can hear NPR, while visible light waves (shorter wavelengths) can be conveniently reflected by simply closing the bathroom door.
This translucent cryolite glass looks the way an iris does: it shows a different hue depending on the color of the background. From this observation we can infer that implanting a light bulb in the back of the eye (so it wouldn’t look dark), would make “blue” irises look yellow.
Here are some other wonders caused by the Tyndall effect, including why the sky is blue.
I love random science walks.

The National Emerging Infectious Diseases Laboratories (NEIDL) is a $200 million research facility built by Boston University. The National Institutes of Health (NIH) agreed to co-fund the project in 2006, but several court injunctions have kept most of its brand-new highly equipped laboratories ridiculously empty.
A recent federal ruling by US District Court Chief Judge Patti Saris determined that the risks to the public associated with the NEIDL are “extremely low, or beyond reasonably foreseeable” and that its current location in Boston’s South End neighborhood poses no greater threat than if it were located in a different suburban or rural area. One more state court lawsuit (which will be decided later this year) stands as the last major obstacle before BSL-3 and BSL-4 research starts taking place at the NEIDL.
Maximum-security research facilities are the best place to pursue our ignorance about how these deadly pathogens work, and they provide the resources to eventually transform scary diseases into manageable annoyances. Unfortunately, a few vocal groups have pushed back against the construction of the NEIDL, claiming that its location would “have a disproportionately adverse effect on minority and low-income population”. This seems completely unfounded. If anyone was at risk, it would be the social contacts of a hypothetically infected lab worker, not the people living close to the facility.
Thirteen pathogens were chosen to be studied in the BSL-3 and BSL-4 facilities within the NEIDL, including the bacterium that causes anthrax, the 1918 flu virus, and the virus that causes SARS. The way I see it, the chances of catching the next swine flu variant coming from China far exceed those of getting infected from one of the pathogens being studied at the NEIDL.
The sooner research starts, the higher the chance that we will keep outbreaks from turning into epidemics.

A mosquito bite is much more sophisticated than a needle prick. The mouthparts that penetrate the skin are surrounded by the labium (lb), which folds and stays outside, the maxillae help make the puncture, the labrum is the tube that sucks up the blood, and the hypopharynx sends saliva down. Once inside the skin, the labrum bends and probes until it finds a capillary (which can take up to two minutes).
Once the capillary is pierced, the mosquito sucks hard—sometimes hard enough to rupture the capillary. The saliva contains anticoagulants, which keeps the blood flowing during the 4-minute-long feeding, and whatever parasites the mosquito was carrying. This video shows the biting process in all its gory details.
This video is freaky
The ice cubes are skin cells, the bendable hook is the mosquito straw, the red lines are the capillaries (you can almost see red blood cells). The images were captured by Valerie Choumet and her colleagues at the Pasteur Institute in Paris by using a microscope to look through a flap of mouse skin. Check out the supplementary section for awesome pictures and videos.
I could talk about mosquito bites all day. Here are a couple of other awesome articles.
I don’t know who that is, but it sure ain’t me.
The International Division of the Pasteur Institute has generously awarded me a grant to carry out a 4-month research internship in its branch located in French Guiana. The goal of the project is to determine the viral biodiversity of multiple species of rodents living in different habitats in the region.
Where I’m going
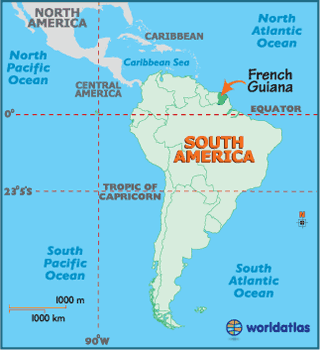
Although French Guiana borders Brazil and Suriname, it is not an independent country, but an overseas region of France. Therefore, it is part of the European Union, the official currency is the euro, and the license plates look European. The population is around 250,000 people, and half of them live in the capital, Cayenne. According to Wikipedia, there is one road that crosses the country parallel to the coast, and the rest is Amazonian jungle.
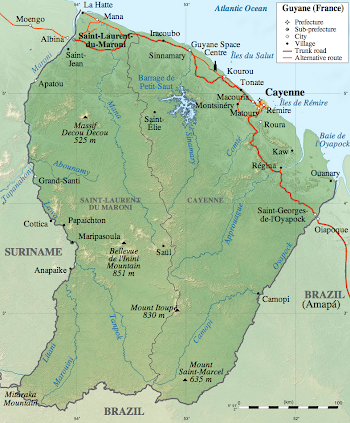
The Pasteur Institute in French Guiana was created in 1940 and it employs around 100 scientists, distributed across 11 different labs. I will be working in the Virus-Host Interactions lab.
What I’m doing
Whenever we urbanize or adapt a terrain for agriculture, we run the risk of disturbing the habitat of the species that live in it. This increases the chances that species that don’t usually interact will come into contact with each other; and raises the possibility that these interactions will help pathogens cross the species barrier, thereby expanding their range. Many different species of rodents live in close proximity to humans, but we don’t know much about the viruses they harbor. The little we know is that three rodent species are the likely reservoirs of some nasty viruses that killed several patients in French Guiana last year.
Metagenomics is a cool bioinformatics technique that can be used to determine all the genetic information that exists in a biological sample. This means that we can go capture rodents all over the country, take a few blood samples, process them and isolate what is likely to viral genetic material. Then, we sequence it and somebody gives me a hard drive full of As, Cs, Gs, and Ts—which is when I come in, because I’m not excited about getting bitten by virus-laden jungle rats.
My job is to determine what sequences are likely to belong to previously known viruses, to identify which ones may correspond to novel viruses (typically 50% of all the generated sequences are completely unknown!), and which ones may be potential human pathogens, and also, to compare the viral families in each sample and see if they correlate better by species or by habitat. Cool stuff.
Stuart Firestein is a neuroscience professor at Columbia University. For the past seven years, he has been teaching a course called Ignorance, where he invites scientists to talk to his students for a couple of hours about the state of their ignorance: what they didn’t know a few years ago and know now, what they would still like to know, why it is important to know it, and how they plan to find out. He’s funny and clever. His TED talk just came out:
Insightful ignorance drives science
Knowing lots of facts doesn’t make you a scientist, you also need to ask good questions. Scientists use facts to ask better questions, insightful questions, questions that probe at the ignorance and expand the frontier of what’s known. Strangely, science is not taught as this exciting, endless pursuit of questions. Rather, it is delivered as lecture-sized bundles of facts that must be memorized to pass the test and have little relevance afterwards. Facts should not be simply accumulated, they should be used to build the next round of questions.
You can look up what we know, but finding out what we don’t know requires having original thoughts. The current educational system doesn’t know how to deal with this requirement. I love the proposal that Stuart shows at the end of his talk about getting rid of tests that ask for the right answer (which Google knows), and replacing them with tests that give the answer, and ask what is the next question (which Google doesn’t know).
Instead of drowning students in pointless facts, teachers should spend most of their time lighting their curiosity. Get them interested enough, and they’ll find out the answer to their questions by themselves.
If you liked his talk, be sure to listen to his interview on the podcast This Week in Virology. He also has a book.
Most bioinformaticians don’t like to do experiments. We love the biological stories, but give us a pipette and we will quickly groan. I have spent the past two years in a virology lab, mainly working on computational problems, and eternally debating if I should plunge into the pool of experimental biology. Up until now, I have only dipped my toes in a few basic experiments, but I have recently decided to stop working on purely computational projects and transition to ones that require experimental skills. Here are my reasons.
I want to learn what’s possible
Computationally, anything I can imagine is possible. Experimentally, I only have a vague notion about what the space of possibility looks like. One of the reasons why I joined an experimental lab was to fill this gap, but I have finally realized that this doesn’t happen by osmosis. I will never understand what techniques are commonly used and which ones are cutting-edge without studying the literature. Also, no amount of reading will teach me how to troubleshoot experiments that don’t work; I need to interact with people that have spent years acquiring the wisdom.
I want to work on more interesting problems
Collaborations between bioinformaticians and biologists typically work like this: 1) biologist approaches bioinformatician with a ton of data and a few questions, 2) bioinformatician spends days analyzing the data, 3) bioinformatician shares findings and waits for biologist to experimentally validate them. This rarely leads to ground-breaking results because computational analyses usually underestimate the biological complexity. Bioinformatics can be used as a hypothesis generation tool, but I find that relegating the experimental component to the end is a mistake. The ability to design and perform any experiment, and the ability to process and analyze any kind of dataset are two ridiculously useful techniques on their own, but when applied together, their usefulness compounds.
"No cilantro, please" Yet I know when it arrives It will be presentHaiku by Krakrs, a member of ihatecilantro.com
The first time I tried cilantro I didn’t realize it; I just thought somebody had emptied a bottle of Old Spice on my pizza in an attempt to poison me. Cilantro tastes like soap to approximately 10% of the people who have had their genotype analyzed by 23andMe. The currently accepted explanation is that those of us who passionately despise cilantro were born with a genetic variant known as a single-nucleotide polymorphism (or SNP, pronounced ‘snip’).
The genome has 3 billion nucleotides (the building blocks, known as A, C, G and T), and 10 million of them are thought to be SNPs. That means that a significant percentage of the population has one letter in a specific location (an A, for example) and everyone else has a different letter at that location. The cilantro SNP is called rs72921001, and apparently, its genomic location lays close to a cluster of olfactory receptor genes that includes OR6A2, the gene most likely to be alerting our brain about the presence of cilantro.
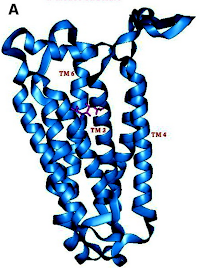 A cartoon representation of the molecular structure of an olfactory receptor. You can think of the red compound snuggling between the helices as the tiniest bit of cilantro.
A cartoon representation of the molecular structure of an olfactory receptor. You can think of the red compound snuggling between the helices as the tiniest bit of cilantro.
When molecules enter the nose, they come into contact with the olfactory sensory neurons located in the upper region of the nasal cavity. Each of these cells expresses only one odorant receptor, and when the right chemical binds to the right receptor, an electrical signal is sent upwards—through the bone—to the olfactory bulb, where it is further relayed by other cells until it reaches the olfactory cortex region of the brain. The OR6A2 receptor gets activated by binding to one of the several aldehydes contained in cilantro.
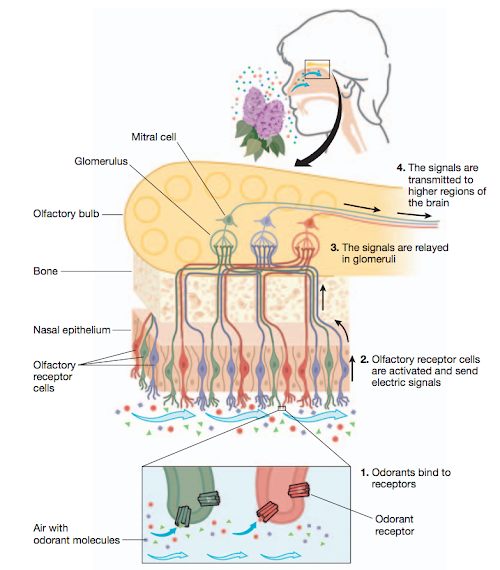 A figure from The scent of life, detailing how smell works.
A figure from The scent of life, detailing how smell works.
The letter you have in the location of the rs72921001 SNP is thought to influence how the OR6A2 receptor perceives the presence of cilantro. If you are among the 10% of humans who have a C instead of an A in that location, when confronted with cilantro, instead of the misleading “Gee, I smell cilantro, lalala, happiness” your brain will warn you of the real danger you face: “I smell the disgusting herb that poisons everything good and loveworthy in this world. Run for your life!”
The science of smell is fascinatingly complex and not entirely understood, but the current hypothesis is that having a C instead of an A in a specific location close to the OR6A2 gene, slightly alters the shape of the receptor molecule encoded by that gene, and allows cilantro molecules to activate receptors that in the rest of the population are only activated by soap/bleach/death molecules.
Whenever I read a paper that describes an experimental technique that I’m not familiar with (which is often, since the lab bench is not where bioinformatic students spend most their time), I compensate by doing background research. Lately, I have been reading a lot about the antiviral effects of interferon-stimulated genes (ISGs), and since John Schoggins is giving a seminar in my university on Monday, I thought I would post about a clever technique that he used in his Nature paper to quantify the impact of ISGs on viral replication.
Interferon-stimulated genes are known to inhibit viral replication, but this effect hasn’t been quantified for all of them. The goal of the Schoggins paper was to determine how individual ISGs change the ability of different viruses to replicate.
The authors used a bicistronic lentiviral vector, which is molecular biology jargon for a piece of DNA that contains two genes that get inserted into the genome of a cell by using a lentivirus virion. They are also known as plasmids or constructs. Besides the two genes, the foreign DNA also contains a bunch of other interesting molecular tools assembled in Frankenstein-like fashion:

LTR stands for long terminal repeat. It is a sequence of nucleotides repeated hundreds of times. Viruses (especially retroviruses, like HIV) use them to insert their genetic material into the genome of their target cell. In this case, instead of inserting a viral genome, they help integrate the plasmid into the genome of a cell.
CMV is one of the enhancer sequences of the human cytomegalovirus genome. It is another viral tool to increase the amount of gene transcription.
IVS-beta is the second intron in the rabbit beta-globin gene (one of the chains that makes up hemoglobin). Adding it to the construct also increases the transcription of the plasmid genes. I haven’t been able to really understand why, but I think it has to do with how it interacts with the splicing machinery.
ISG is the sequence for one of the 300+ known interferon-stimulated genes. Inserting it after the two previous elements ensures that it will be highly transcribed.
EMCV IRES is the internal ribosomal entry site of the encephalomyocarditis virus. This is another page taken from the virus book: it is a nucleotide sequence that allows ribosomes to bind to the middle of the messenger RNA molecule, instead of the more traditional 5’ end. Viruses use it to synthesize their proteins when the host translational machinery is not supposed to be working. In the plasmid above, it ensures that the RFP gene is translated in a proportional amount to the ISG gene.
RFP stands for red fluorescent protein. Using it as the second protein of the plasmid makes it easy to identify which cells have incorporated the plasmid into their genome, and are therefore, also expressing the ISG of interest.
The viruses used in the experiment were modified so they would express GFP (green fluorescent protein). That way, the amount of viral replication can be quantified by measuring the amount of red fluorescence (from cells that incorporated the plasmid) and green fluorescence (from cells that were also infected with the virus). Each ISG construct has a specific ability to reduce viral replication. Surprisingly, a number of ISGs actually enhance viral replication (the circles that are above 100% in the graph below), although the reasons for this are not yet clear.
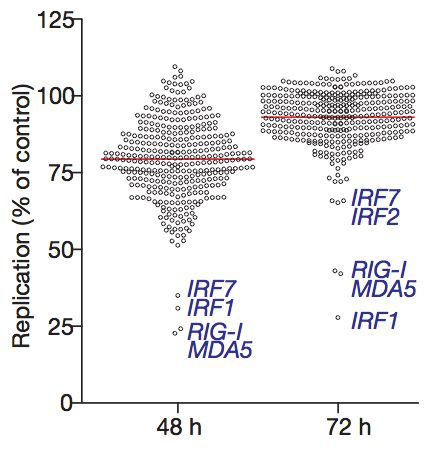
Clever molecular techniques like this one are the best tool that we have to understand what is really going on inside our cells. By studying them, I hope to learn enough to eventually design my own useful experiments.
I gave a presentation today in the Microbiology departmental retreat at Boston University, so I though I’d share it.

The goal of the research project I have been working on for the past two years, as part of my Bioinformatics PhD at Boston University, is to use blood samples from infected patients to determine which virus is causing the infection.
Traditional diagnostic methods, like detection by ELISA, are only effective after the virus has had enough time to replicate in the blood of the patient. However, for some viral infections, when this happens it is already too late for the treatment to be effective.
The approach that we have taken in my lab is to measure the transcriptional changes that take place in the peripheral blood cells, and identify patterns of expression that are unique for each type of infection. This indirect way of viral detection has the potential to reduce diagnosis time by several days, since it is known that these transcriptional changes precede the appearance of virus particles in the blood, and we believe that they are specific enough to discriminate between viruses.
To test our hypothesis, we sequenced the RNA from PBMCs that were extracted from two groups of monkeys; one infected with Lassa virus, and the other with Marburg virus.
After comparing the expression of the samples extracted 3 days post-infection with those taken before infection, we identified multiple genes that showed strong changes in expression. Most of these genes play important roles in innate immunity pathways, so we expected them to be highly expressed in both infections.
What we found most surprising was the number of genes that showed different patterns of expression depending on what virus was causing the infection. Using only the expression patterns of a handful of genes, we were able to correctly classify unknown samples into three groups: uninfected, infected with Lassa virus, and infected with Marburg virus.
Our hope is that these results will be integrated in a diagnostic kit to easily identify Lassa and Marburg infected patients in areas where these diseases are common, but there is still a long way to go.
Here is the presentation I gave today.
Lately I have been summarizing every paper I read on a single piece of paper. Before I started doing that I would invest a lot of time reading a paper, and two months later it would completely disappear from my brain. The only thing I would remember is if I had found it interesting or not (not the most important information when trying to compile a bibliography). Limiting myself to a single piece of paper ensures that I only record what is important, and the fact that the notes are handwritten lets me triple underscore and use all the curvy arrows I need to make important ideas stand out. My two-month recall is now pretty decent.
I’m going to a seminar tomorrow presented by Tim Lu, one of the authors of a Nature Biotechnology paper titled Synthetic circuits integrating logic and memory in living cells. I decided to read the paper and write up a quick summary.

The paper describes a new technique to arrange the genome of a bacterium (E. coli) and turn it into the biological equivalent of an electronic circuit. This approach is much more scalable than previous techniques because using it to build complex circuits like an XOR gate doesn’t require joining universal gates together, which means that instead of assembling this madness:

you can assemble the XOR gate directly:
(By the way, a gate is just a way to convert two inputs into one output. Inputs and outputs can be either 1s or 0s. The type of gate (AND, OR, XOR, etc.) describes the rule that is used to make this conversion. For example, check out the table rule for the AND gate)
Stinginess is important because you can use as many components as you can fit in a circuit boards, but cells are much more puritan about the number of resources they allow.
The paper’s magic sauce is in the use of inducible recombinases—recombinases that become active when their inducer (a riboregulator) is added to the media. An activated recombinase can bind specific regions of DNA and flip their orientation, which in turn switches on or off the expression of their downstream gene.
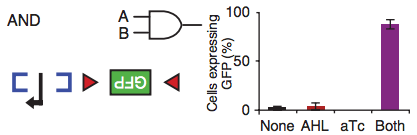
For example, the blue brackets in the figure above represent the DNA binding site of a recombinase that, once activated by its inducer (input A), flips that region of DNA, making the promoter (black arrow) accessible to the RNA polymerase. But the RNA polymerase has nothing to transcribe unless the region between the red triangles is also flipped by a different recombinase. When this recombinase gets activated by its inducer (input B), the Green Fluorescent Protein gene can be transcribed and the cell starts making enough GFPs to turn the bacterium into a swimming Christmas tree. As the barplot shows, when both inputs are not present, the amount of GFP that is made is minimal (no biological circuit will ever be perfect).
(Just to clarify, GFP is an example, any gene can be integrated into this type of circuit. It’s just that a glowing bacterium is easier to detect than one that expresses the blond-hair gene)
That is just one possibility out of the 16 that appear in Figure 2 (check out the especially clever technique used to build the XOR and XNOR gates).
Because the recombinases modify the information that is stored in the bacterial chromosome, after a bacterium divides, the two daughter cells have the same genetic information, which means that next generations of bacteria will keep synthesizing GFP for centuries (days, in bacterium-years). DNA is what Nature uses as a hard drive, which means that coming up with clever ways to store information in it will bring us ever closer to genetically engineering the bionic pets we have all been dreaming about.
The paper is blissfully short, and worth a read if you think synthetic biology is cool.